- Volumes 96-107 (2025)
-
Volumes 84-95 (2024)
-
Volume 95
Pages 1-392 (December 2024)
-
Volume 94
Pages 1-400 (November 2024)
-
Volume 93
Pages 1-376 (October 2024)
-
Volume 92
Pages 1-316 (September 2024)
-
Volume 91
Pages 1-378 (August 2024)
-
Volume 90
Pages 1-580 (July 2024)
-
Volume 89
Pages 1-278 (June 2024)
-
Volume 88
Pages 1-350 (May 2024)
-
Volume 87
Pages 1-338 (April 2024)
-
Volume 86
Pages 1-312 (March 2024)
-
Volume 85
Pages 1-334 (February 2024)
-
Volume 84
Pages 1-308 (January 2024)
-
Volume 95
-
Volumes 72-83 (2023)
-
Volume 83
Pages 1-258 (December 2023)
-
Volume 82
Pages 1-204 (November 2023)
-
Volume 81
Pages 1-188 (October 2023)
-
Volume 80
Pages 1-202 (September 2023)
-
Volume 79
Pages 1-172 (August 2023)
-
Volume 78
Pages 1-146 (July 2023)
-
Volume 77
Pages 1-152 (June 2023)
-
Volume 76
Pages 1-176 (May 2023)
-
Volume 75
Pages 1-228 (April 2023)
-
Volume 74
Pages 1-200 (March 2023)
-
Volume 73
Pages 1-138 (February 2023)
-
Volume 72
Pages 1-144 (January 2023)
-
Volume 83
-
Volumes 60-71 (2022)
-
Volume 71
Pages 1-108 (December 2022)
-
Volume 70
Pages 1-106 (November 2022)
-
Volume 69
Pages 1-122 (October 2022)
-
Volume 68
Pages 1-124 (September 2022)
-
Volume 67
Pages 1-102 (August 2022)
-
Volume 66
Pages 1-112 (July 2022)
-
Volume 65
Pages 1-138 (June 2022)
-
Volume 64
Pages 1-186 (May 2022)
-
Volume 63
Pages 1-124 (April 2022)
-
Volume 62
Pages 1-104 (March 2022)
-
Volume 61
Pages 1-120 (February 2022)
-
Volume 60
Pages 1-124 (January 2022)
-
Volume 71
- Volumes 54-59 (2021)
- Volumes 48-53 (2020)
- Volumes 42-47 (2019)
- Volumes 36-41 (2018)
- Volumes 30-35 (2017)
- Volumes 24-29 (2016)
- Volumes 18-23 (2015)
- Volumes 12-17 (2014)
- Volume 11 (2013)
- Volume 10 (2012)
- Volume 9 (2011)
- Volume 8 (2010)
- Volume 7 (2009)
- Volume 6 (2008)
- Volume 5 (2007)
- Volume 4 (2006)
- Volume 3 (2005)
- Volume 2 (2004)
- Volume 1 (2003)
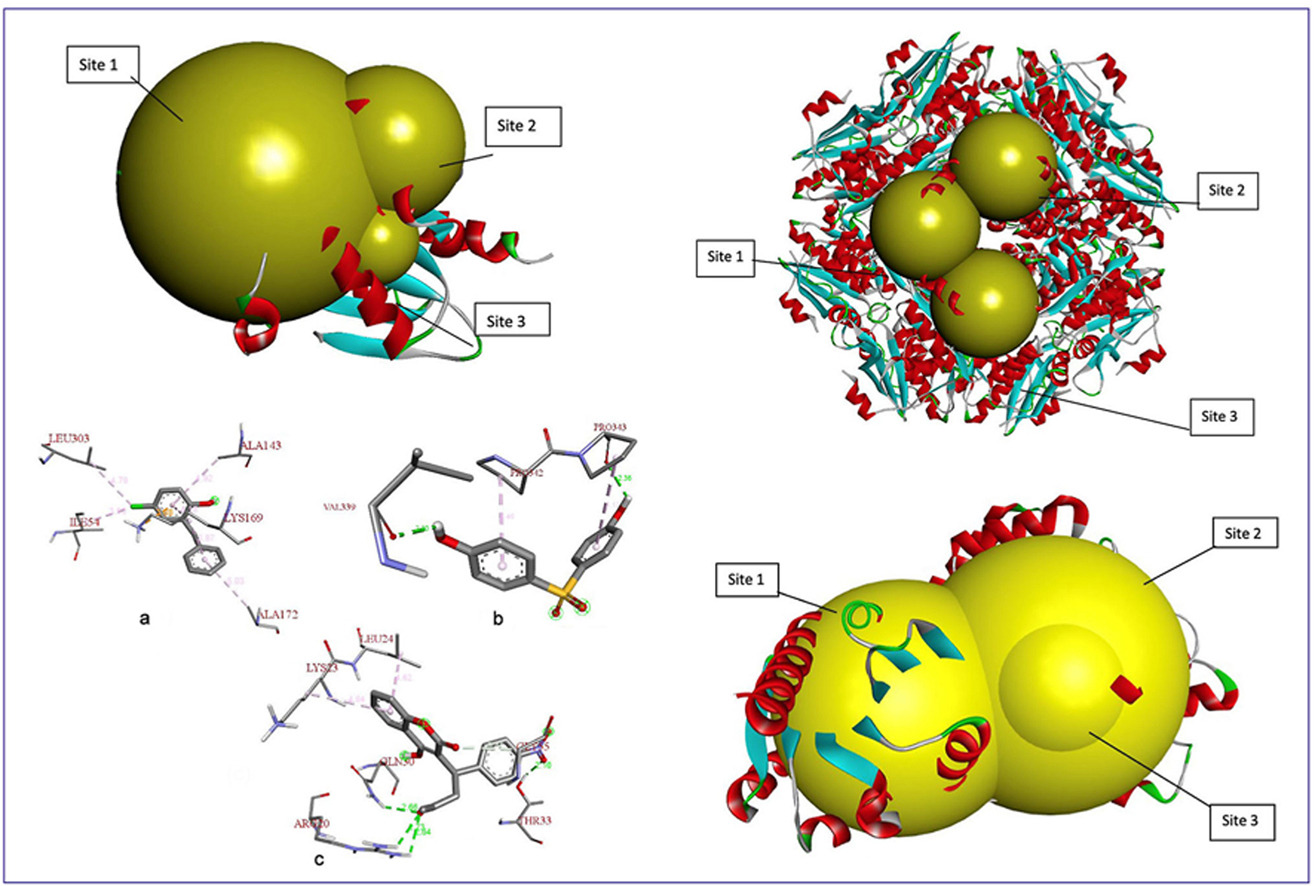
• Aeromonas caviae shows natural potential in biodegradation due to diverse enzymatic pathways.
• Docking shows synergy of beta-ketoadipate enol-lactone hydrolase and muconate cycloisomerase in phenol breakdown.
• Ligand-protein docking revealed effective interactions in the hybrid biodegradation enzyme model.
• Bioengineered hybrid enzyme showed better docking scores and structural similarity to known homologs.
• Enzyme bioengineering enhanced the biodegradation potential of marine-derived phenolic compounds.
Toxic aromatic compounds are widespread environmental pollutants posing significant ecological and health risks due to their persistence and toxicity. Aeromonas caviae is a promising microbial candidate for biodegradation owing to its diverse enzymatic arsenal. This study investigates the cooperative roles of two key enzymes from A. caviae, beta-ketoadipate enol-lactone hydrolase, which is involved in ring-opening hydrolysis, and muconate cycloisomerase, which catalyzes the isomerization of muconate intermediates, in degrading 15 selected toxic compounds using computational methods. Enzyme stability analysis via ExPASy ProtParam indicated an instability index below 40, confirming structural stability. Homology models were constructed and validated with high-quality scores. Molecular docking revealed Acenocoumarol as the compound with the highest binding affinity (−7.8 kcal/mol). Protein-ligand interaction analysis identified key residues involved in Pi-Pi and hydrogen bonding interactions critical for catalysis. Furthermore, a bioengineered hybrid enzyme model demonstrated improved binding precision and structural similarity to homologous proteins. These findings highlight the potential application of A. caviae enzymes in the bioremediation of toxic pollutants. Future experimental validation and enzyme engineering could further enhance their catalytic efficiency for sustainable environmental detoxification.